Laboratory testing for 2019 novel coronavirus
(2019-nCoV ) in suspected human cases.
Specimen collection and shipment
Samples to be collected (see Table 1 for details on samplecollection and storage):
1-Respiratory material
(nasopharyngeal and oropharyngeal swab in ambulatory patients and sputum (if produced) and/or endotracheal aspirate or bronchoalveolar lavage in patients with more severe respiratory disease)
2- Serum for serological testing, acute sample and convalescent sample
(this is additional to respiratory materials and can support the identification of the true agent, once serologic assay is available)Modifiable with information on whether upper or lower respiratory material is better for coronavirus detection.
A single negative test result, particularly if this is from an upper respiratory tract specimen, does not exclude infection.
Repeat sampling and testing, lower respiratory specimen is strongly recommended in severe or progressive disease.
Apositive alternate pathogen does not necessarily rule out either, as little is yet known about the role of coinfections.
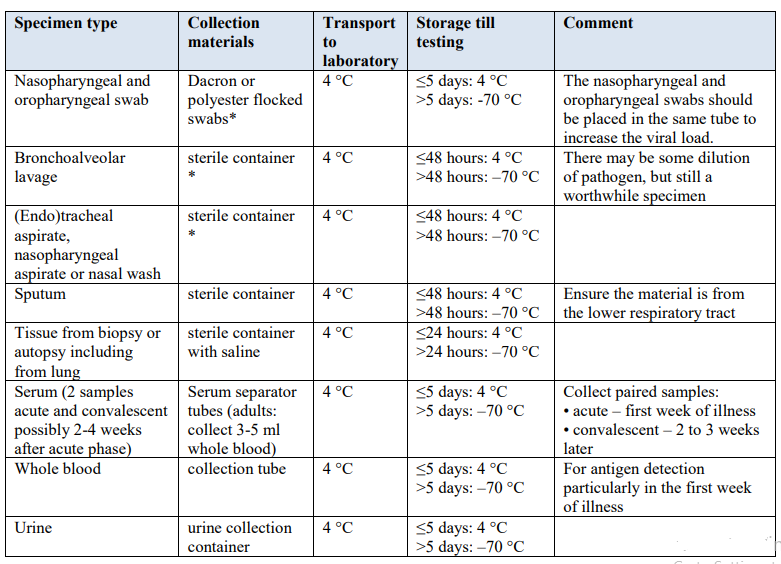
Table 1. Specimens to be collected from symptomatic patients
Guidance on specimen collection
----------------
Testing of 2019-nCoV in reference
laboratories
2019-nCoV testing for patients that meet the suspected
case definition
Patients that meet the case definition for suspected 2019-
nCoV should be screened for the virus with PCR . If case management requires, screen also for other
common causes of respiratory illness according to local
guidelines (1,5,7). As coinfections can occur, all patients
that meet the case definition should be tested for 2019-nCoV
regardless of whether a conventional respiratory pathogen is
found. If testing does not occur in an expert/reference
laboratory it is encouraged to send the sample for
confirmation to a regional, national or international
reference laboratory with pan-coronavirus or specific 2019-
nCoV detection capacity. WHO can assist Member States to
identify laboratories able to provide this support.
Nucleic acid amplification tests for 2019-nCoV
As sequence information from the 2019-nCoV has recently
been made available, PCR assays can be designed to detect
these sequences. PCR assay design optimization can be a
complicated process, and a useful option is to contact the
experienced laboratories publicizing their assays and request
access to their assay chemistries.
Laboratories may desire to use a pan-coronavirus assay for
amplification followed by sequencing of amplicons from
non-conserved regions for characterization and
confirmation. The importance of the need for confirmation
of results of testing with pan-coronavirus primers is
underscored by the fact that four human coronaviruses
(HcoVs) are endemic globally: HCoV-229E, HCoV-NL63,
HCoV-HKU1 as well as HCoV-OC43. The latter two are
betacoronaviruses. Two other betacoronaviruses that cause
zoonotic infection in humans are MERS-CoV, acquired by
contact with dromedary camels and SARS arising from
civets and cave-dwelling horseshoe bats.
Alternatively, amplification and detection of 2019-nCoV
specific sequences can be diagnostic without the necessity
for further sequencing. In the case of surprising findings or
for less-experienced laboratories, external assistance should Once specific NAAT assays are developed and validated,
confirmation of cases of the novel virus infection will be
based on specific detection of unique sequences of viral
nucleic acid by reverse-transcriptase polymerase chain reaction (RT-PCR). Alternative NAAT techniques with
advantages of greater speed or simplicity of use may also
become available.
Serological testing
Serological testing may be useful to confirm immunologic
response to a pathogen from a specific viral group, e.g.
coronavirus. Best results from serologic testing requires the
collection of paired serum samples (in the acute and
convalescent phase) from cases under investigation.
(details below in this book)
Chemist / Aly Alaraby











هناك تعليق واحد:
Listen...
What I'm going to tell you may sound a little weird, and maybe even a little "out there..."
BUT what if you could simply hit "Play" to listen to a short, "magical tone"...
And magically attract MORE MONEY into your LIFE??
And I'm really talking about thousands... even MILLIONS of DOLLARS!!
Think it's too EASY?? Think this couldn't possibly be for REAL??
Well then, I'll be the one to tell you the news.
Many times the greatest miracles in life are the SIMPLEST!!
In fact, I will provide you with PROOF by letting you listen to a real-life "miracle wealth building tone" I've synthesized...
You simply click "Play" and watch how money starts piling up around you.. starting almost INSTANTLY..
GO here now to enjoy the magical "Miracle Abundance Tone" - as my gift to you!!
إرسال تعليق